Working in R interactively
To see the versions of R that are installed as modules, type "module spider r". Note that "module spider R" is not that helpful.
module load r
Or use either
module load r/3.5.0
module load r/3.5.1s
to specify a specific version of R.
Type "R" to begin an interactive session in R ("r" does not work). This is one way to use R, because you are working on a single node, but you can play around in it. (Preferably when you're not on a log-in node, else you slow down the log-in node for everybody else.)
You will need to install R packages into some directory available to you. When you first install a package with R, it will ask you if you would like to install in a directory within your /home folder. You could install these in any directory, but you would need to add that path using libPaths() so that R knows where to look. You can do this at the beginning of your scripts, or add it to your .Rprofile instead.
Troubleshooting loading R packages (Jason would you move this to the troubleshooting page? Thx! LCM)
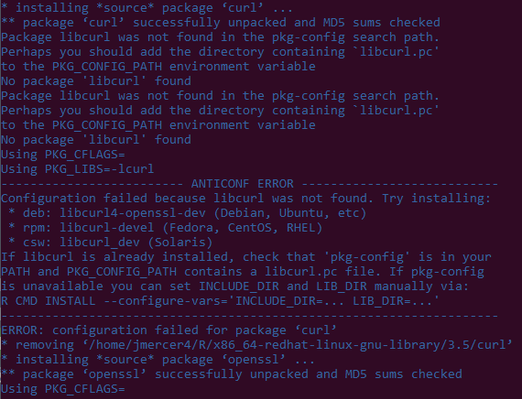
I'm getting the following error when trying to install the curl package from inside R (using load module r/3.5.1s) , and don't know how to interpret it, or how to fix it:
I've tried using different CRAN mirrors, but get the same result.
LCM - On Moran this showed you the R versions available, but on Teton it's not very helpful because it shows every module with the letter r in it.
- I loaded this, but I am unclear as to what I am looking at. It appears to be a bunch of helper functions, or something, that aren't directly associated with a "typical" R experience. Jason Mercer I will flesh this out but until then, here's info on spider: https://arccwiki.uwyo.edu/index.php/Software:_Lmod
- How do I load an R package? Is that even possible?
- Are there best practices for testing code before sending it to Teton (to make sure the code is actually doing what I think it should be doing)? I'd like to not take up more resources than I need.
- What are the maximum number of nodes I can use for parallel computing?
- Who do I talk to about adding Stan to a module?
Running an R script non-interactively
The way that you will want to run R scripts is to use sbatch commands that will send your R script through the SLURM job scheduler. Before you can run an R script non-interactively, you will need to install all the packages your script uses in a path that R can reach, as described above. Then, to run your script non-interactively as a slurm job, you should write a slurm/sbatch wrapper for it. The sbatch script will need to include a "module load R" call; you can specify the R version if you like. Then, you can load your R packages within the R script itself. The syntax for actually running the script within the slurm wrapper has two options:
Rscript yourscript.R
R CMD BATCH
(Is this right or is only the Rscript command proper for using within a slurm wrapper?)
Troubleshooting R (Jason would you move this too? Thx! LCM)
Jason (I need help with this issue – wish I knew how to issue track):
To get R running, I used the following command before starting R:
module load r/3.5.1s
I used that, rather that module spider ..., because I don't understand what spider is doing (I find its documentation either lacking or cryptic), though I recognize that there are recommendations that I use spider to load modules. However, what I was doing was working well for me, because I was able to install packages that were maintained across sessions, which is exactly what I want. However, I was continuously encountering the following error:
During startup - Warning messages: 1: Setting LC_CTYPE failed, using "C" 2: Setting LC_TIME failed, using "C" 3: Setting LC_MESSAGES failed, using "C" 4: Setting LC_MONETARY failed, using "C" 5: Setting LC_PAPER failed, using "C" 6: Setting LC_MEASUREMENT failed, using "C"
I played around with some toy problems, and the error didn't seem to be impacting the results I'd expected, but I'd not been exhaustive in my investigation, and the constant display of this error made me nervous, especially since it came up a number of times during the installation of packages. After looking around a bit (e.g., https://stackoverflow.com/q/9689104), I thought maybe I could modify my bash profile to ensure the right encoding was being used (I think that's the right concept; e.g., https://stackoverflow.com/q/30264526). To modify my profile, and get rid of the errors, I did the following:
- Logged on to Teton
Logged off the login node:
srun --pty --account="<project>" -t 0-01:00 /bin/bash- Checked that
.bash_profileexists:ls -a- Hopefully you see the bash profile file.
- Edited the profile file with Vi/Vim:
vi .bash_profile - Added the following line to the bottom of the file:
export LANG=en_US.UTF-8- To be clear, this line of code should be placed above all the `~` values in the file.
- Exited Vim and saved the file: <ctrl>+c then type
:wq! - To make sure my profile loaded properly, I closed out of Terminal (or bash, or whatever) and logged back on to Teton. (I've noticed that new aliases I make need a fresh terminal, but the same concept may not apply here.)
- Logged off the login node.
- Load R:
module load r/3.5.1s(version 3.5.1s was the most current on Teton as of writing this how-to) - Open R:
R - Check for the errors from above, which hopefully don't pop up.
My current problem is that I can't install all the packages I'd like. For example, openssl, which is part of the tidyverse and devtools packages, will not load. I really have no idea why this and other packages aren't loading, but it may be because we are currently on R version 3.5.2. This could mean it is a versioning issue, but that's just one hypothesis.